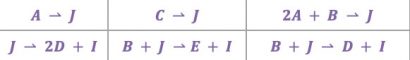Imagine the case of a scientist investigating a certain reaction in one of the several robotic devices in the market. He can easily collect a lot of data. How can these data be used to understand the stoichiometry of the reactions taking place? He/she knows the stoichiometry of the primary reaction, of course. The collected composition data are at different time instants during a set of experiments. She/he has developed a DRSM model of each of the measures species. The measured and modeled species include the known reactants and desired products and a lot more. These are the expected and unexpected intermediates and byproducts. And you are wondering what the true stoichiometric description of the complex reaction network is. Here is where our stoichiometric discovery methodology can help.
With the DRSM models at hand for all the species measured in the reaction mixture, one can calculate the number of linearly independent reactions taking place. Furthermore, one can then proceed to discover the active stoichiometry. This is achieved by Target Factor Analysis (TFA) (Bonvin and Rippin, 1990), a methodology not utilized fully until recently, through the help of the DRSM models.
Our stoichiometric discovery methodology, using matrix operations and statistical tests, can identify the most significant of the true reaction stoichiometries from a set of candidates. The outline of the methodology is:
- Development of DRSM models for all measured species
- Differentiation, with the time, of all concentration models to calculate the rate of appearance or disappearance of each measured species
- Singular value decomposition of a matrix with the above rate data
- Statistical Estimation of the number of linearly independent reactions
- Testing of candidate stoichiometries to see if they agree with the data
Challenging Example
This methodology has been successfully tested against the following challenging example, postulated to us a few years ago by our Pfizer collaborators. Most of the major correct reactions were successfully identified by the above methodology. The example consists of the following eight reactions among the ten measured species. With some assumed kinetic models, a set of experiments were simulated, and measurement error was added. The composition data was passed to us, and we were asked to identify the correct stoichiometries from a set of candidates.
The following set of eight reactions were used in the simulation program. Our methodology correctly identified six of the eight reactions. It only missed the last two reactions which had very small reaction rates, which were obfuscated by the substantial error added.
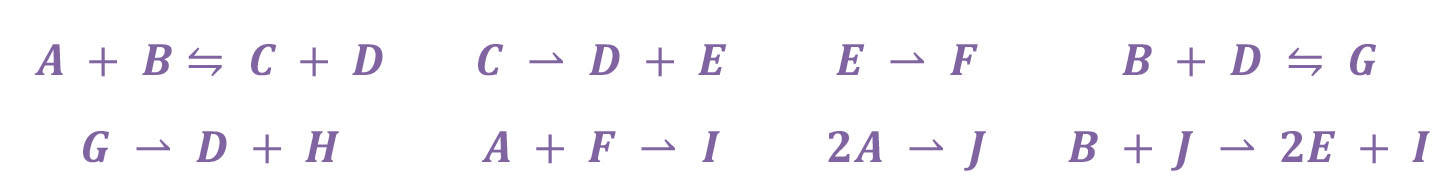
To the above eight true reactions and the six false, below, were added to the candidate stoichiometries. The stoichiometry discovery algorithm rejected all six of the false reactions.